Learn how to install R packages from command line using the install.packages() function, CRAN mirrors, and GitHub repositories. This complete guide covers package management in R, including installing, loading, and removing packages from terminal.
Whether you need to install packages from CRAN, GitHub, or local files, this tutorial shows you the exact R commands for command line package installation. We'll also cover CRAN mirror selection, package dependencies, masking conflicts, and troubleshooting common installation errors.
What is R Package Management? Complete Guide
Simply put, an R package consists of a series of R functions, such as datasets, support files, and compiled code, packed compact and well-defined.
These packages are just compressed files that must be unzipped and placed in the right location on your machine before you can use them in R.
This entire process happens automatically and does not require user intervention. Aside from the software itself, the R installation file includes about 30 default or recommended packages, where about seven are loaded into memory immediately once R is launched.
These packages are mounted in a designated folder on your computer and are used for a broad range of computational tasks, such as data management and statistical analysis.
Nevertheless, because R is an open-source programming language, many user-contributed packages are available for various purposes and are publicly accessible to anyone.
These packages can be accessed from the CRAN website or R's repositories. The following segment provides further specifics regarding how to download and install the user-contributed R packages.
Launch R and execute the following command in the R console:
search()And the output:
 R console showing the search() command output
R console showing the search() command output
The R output above (second and third line) displays a collection of search path items. The numbers in square brackets show the positional index of the unit immediately to the right, e.g., the number [5] indicates that the fifth item in the list, respectively, is the grDevices package.
You can observe that not all packages have a position number [x] assigned to them, but as long as you can count them, you can figure out the respective position yourself.
The above output may appear slightly different on your R system. For instance, on other displays, the R console window will be sized automatically to fit the supported resolution, especially in width. As a result, the R console will be scaled to match it.
Another reason might be that you may have other packages loaded in the system, either because you installed them yourself or because the R team has included (or removed) regular packages in newer versions of R.
Next, let’s have a look at some of the most important options in the search() output command shown above:
The GlobalEnv entry is often located in the first position [1] of the search route and is not an R package. The GlobalEnv stands for Global Environment and represents the location where freshly created R objects are placed in the memory.
The package package:base is always positioned last, and unlike the other packages, package:base cannot be removed.
If you are using RStudio, you will see an additional entry here, respectively tools:rstudio, in addition to the output above.
Remember that the 30 default R packages are not all loaded in the memory when R is launched. You can use the function library in R to load any of these or any other package installed but not yet loaded in R memory.
For instance, the package MASS is one of the 30 packages installed but not loaded into the memory. I will use the command library to load it:
library(MASS)And the output:
 Loading the MASS package using the library() command
Loading the MASS package using the library() command
As you can see, the package:MASS is loaded in the position [2] once installed, and all the other packages are pushed to one position higher.
It is important to remember that the location [x] of each package in the search path is significant as it establishes the priority for replication of functions.
When we load packages into R memory, we ensure the functions in these packages are available throughout the session. Remember, memory management plays a crucial role in programming.
Packages no longer necessary in a session can be excluded from the search route. To unload a package from memory, you need to use the command detach as follows:
detach(package:MASS)You can also remove a package from memory by specifying its position number, as shown in the picture below:
detach(pos=2)You will remove the stats package from the R search path if you run the previous commands. Once removed, you will receive an error message if you want to use the functions in this package.
If you’re looking for more details, check the help page of detach for more information. You can load the package anytime you want without any adverse effects on your system using the command below:
library(stats)Note: As mentioned before, the package:base and GlobalEnv packages cannot be removed.
An essential thing to remember is that packages that you load manually using the library command will be automatically detached when you quit R and not be reloaded when you start another R session.
And one more thing. You may have heard about user-contributed R packages. If not, you should know that these packages are developed by R users worldwide and are entirely free.
The user-contributed packages are available on the CRAN project website and are quite a number for any possible scenario you may need. Go have a look.
There are over 14,000 community packages ready to install and use in R at the time of writing this tutorial, though the number increases by the day. The user-contributed packages aim to reduce the complexity of numerous commands required to accomplish specific tasks in R.
How To Install R Packages
When we choose to use a function or dataset from a user-contributed package in R, we must follow two basic steps:
1. Install R packages by executing the install.packages() function.
If you downloaded the package manually from the Internet, you can install it directly from the respective .zip or .tar file.
Let’s install the MySQL package for R this way using the command:
install.packages("RMySQL")R will most likely show the following output message in your terminal:
# - Please select a CRAN mirror for use in this session -
This message means R cannot find the RMySQL package in its repository, and we must install a mirror first.
If you downloaded the package manually from the Internet, you can install it directly from the respective .zip or .tar file by simply introducing the path to the file location on your computer between the “” in the command above.
2. Install R packages using CRAN mirrors.
For instance, when installing the R RMySQL package, you will be asked to select a CRAN mirror from where the package is downloaded.
You can get a repository list window or a text menu with a few choices. But if it doesn’t appear, you can still choose the mirror from which to import the packages using the repositories parameter repos=, and after doing so, R will no longer bother you to select a CRAN mirror.
Here is an example of using the US mirror to get the RMySQL package in my R system:
install.packages('RMySQL', repos='http://cran.us.r-project.org')And the command output:
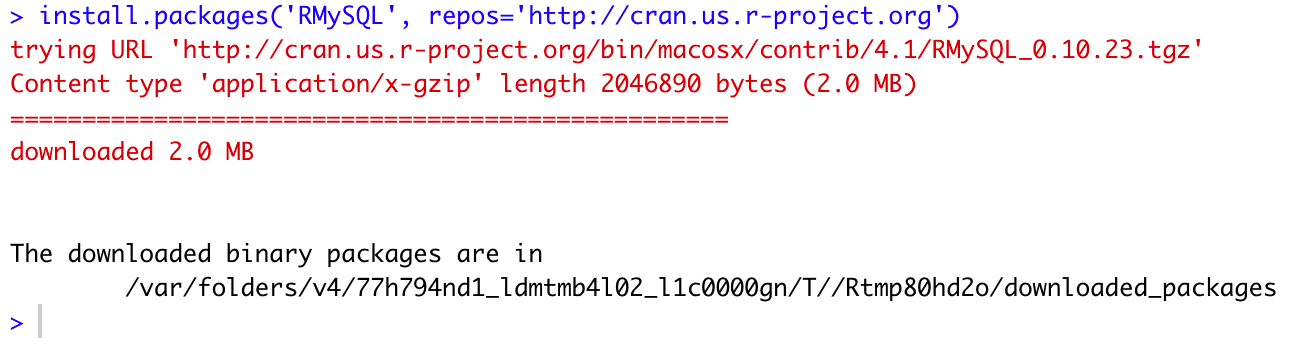 Installing RMySQL package from CRAN mirror
Installing RMySQL package from CRAN mirror
Here, you can find the list of all available R mirrors in various geographical locations. You should select the nearest CRAN mirror to your place, especially if you have a slow Internet connection.
Finally, it is essential to remember that a package that depends on another cannot be detached from the system.
How To Install R Packages From Source
Installing R packages from source code gives you access to the latest development versions and allows you to install packages not available on CRAN. This method is particularly useful when you need specific features or bug fixes that haven't been released yet.
To install an R package from source, download the package source file (usually a .tar.gz file) from CRAN or the package repository. Then use the following command:
install.packages("/path/to/package_version.tar.gz", repos = NULL, type = "source")For example, if you downloaded the dplyr source package:
install.packages("~/Downloads/dplyr_1.0.7.tar.gz", repos = NULL, type = "source")The repos = NULL parameter tells R that you're installing from a local file, not a repository. The type = "source" parameter specifies that this is a source installation.
Installing from Source on Windows
On Windows, installing from source requires Rtools, which provides the necessary build tools. After installing Rtools:
install.packages("packagename", type = "source")Installing from Source on macOS
On macOS, you may need Xcode Command Line Tools for compiling packages from source:
xcode-select --installThen install the R package from source:
install.packages("packagename", type = "source")Installing from Source on Linux
Linux systems typically have the necessary build tools installed. If you encounter compilation errors, install the development packages:
sudo apt-get install build-essential gfortranInstalling from source is particularly useful when working with packages that have C, C++, or Fortran code, as it compiles the code specifically for your system architecture.
Where Are R Packages Installed? (Windows, Mac, Linux)
Depending on the operating system you are using (Windows, macOS, Linux/UNIX) or your user privileges, the location of the installed R packages may differ, as well as the access to the R package installation folder.
To find out the patch where R is storing its packages, type in R the following command:
.libPaths()Typically, on a Windows machine, the R packages will be located in the “C:\Program Files\R” folder.
On a macOS machine, the R packages are typically installed in the “/Library/Frameworks/R.framework/Resources/library” folder.
 Checking R package installation path using .libPaths()
Checking R package installation path using .libPaths()
If you prefer a custom location to install the R packages, you will need to define it in the .Rprofile. For instance, on a Mac computer, we can instruct R to install R packages at a custom location using:
.libPaths( "/Users/tex/lib/R" )The .Rprofile file on Windows is typically located in the C:\Program Files\R\R-...\etc folder, but you can specify a custom location during the R installation wizard.
R will remember the new path and install the packages at this location from now on.
How To Install R Packages Using RScript
RScript allows you to install R packages directly from the command line without opening the R console. This method is particularly useful for automation, scripting, and server environments.
To install an R package using RScript from your terminal, use the following command:
Rscript -e "install.packages('packagename', repos='http://cran.us.r-project.org')"For example, to install the ggplot2 package using RScript:
Rscript -e "install.packages('ggplot2', repos='http://cran.us.r-project.org')"The -e flag tells RScript to execute the R code provided in quotes. The repos parameter specifies the CRAN mirror to use, preventing the interactive mirror selection prompt.
Installing Multiple Packages with RScript
You can install multiple R packages in a single RScript command by using the c() function:
Rscript -e "install.packages(c('dplyr', 'tidyr', 'ggplot2'), repos='http://cran.us.r-project.org')"Running RScript Files for Package Installation
For more complex package management tasks, you can create an R script file and execute it with RScript:
# install_packages.R
packages <- c("dplyr", "ggplot2", "tidyr", "readr")
install.packages(packages, repos="http://cran.us.r-project.org")Then run the script from terminal:
Rscript install_packages.RThis approach is ideal for setting up reproducible R environments on new systems or continuous integration pipelines.
Install R Packages on Linux Command Line
Installing R packages on Linux command line follows the same principles as other operating systems, but there are some Linux-specific considerations for package dependencies and permissions.
Basic Linux Installation
On Linux, you can install R packages from the terminal using RScript:
Rscript -e "install.packages('packagename', repos='http://cran.us.r-project.org')"Linux System Dependencies
Many R packages require system-level libraries on Linux. For example, the curl package requires libcurl-dev, and xml2 requires libxml2-dev.
On Ubuntu/Debian, install system dependencies first:
sudo apt-get install libcurl4-openssl-dev libxml2-dev libssl-devOn Fedora/RHEL/CentOS:
sudo yum install libcurl-devel libxml2-devel openssl-develThen install the R package:
Rscript -e "install.packages('curl', repos='http://cran.us.r-project.org')"Linux User vs System-Wide Installation
By default, R installs packages to your user library. To install packages system-wide on Linux (requires sudo privileges):
sudo Rscript -e "install.packages('packagename', repos='http://cran.us.r-project.org', lib='/usr/local/lib/R/site-library')"Check your library paths on Linux:
Rscript -e ".libPaths()"Installing R Packages from Linux Package Managers
Some Linux distributions provide R packages through their package managers. On Ubuntu/Debian:
sudo apt-get install r-cran-packagenameHowever, CRAN usually has more recent versions, so using install.packages() or RScript is generally recommended.
How To Install R Packages From GitHub
Some R packages developed by the R community are located on GitHub. To install R packages from GitHub, we will need to install the devtools package in R first. To do this, type in the R console the following command:
install.packages("devtools") Installing devtools package to enable GitHub installations
Installing devtools package to enable GitHub installations
The devtool package and several dependencies are now installed in your system. However, devtool is not loaded in R memory yet; therefore, we need to instruct R to do so using the following command:
library(devtools)To install R packages from GitHub, head over to GitHub and take note of the package author and package name.
In this example, I will install Allison Horst’s palmerpenguins package by using the install_github function.
install_github("allisonhorst/palmerpenguins")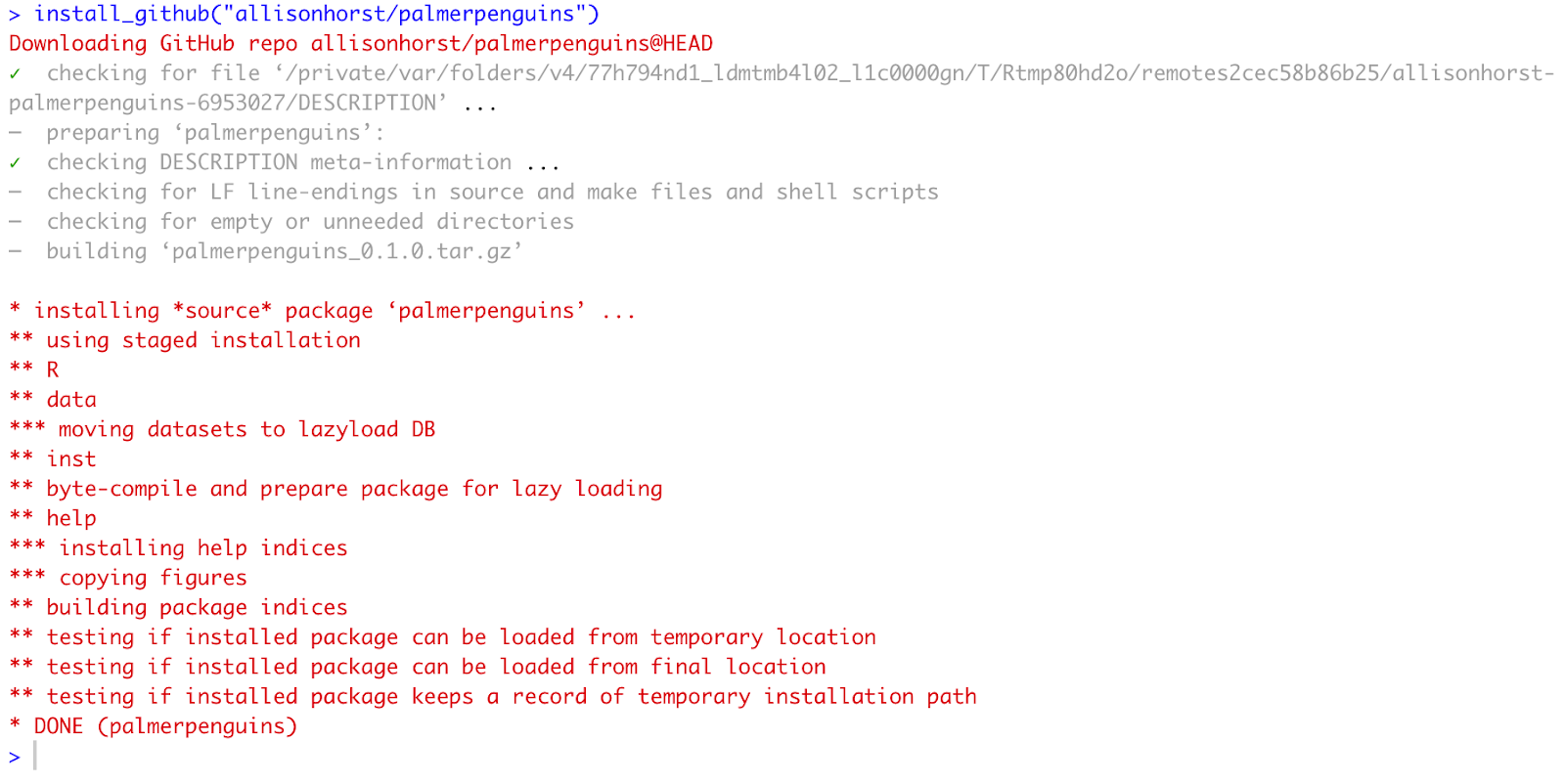 Installing palmerpenguins package from GitHub
Installing palmerpenguins package from GitHub
As you can see, the palmerpenguins is now listed in the Packages tab in R.
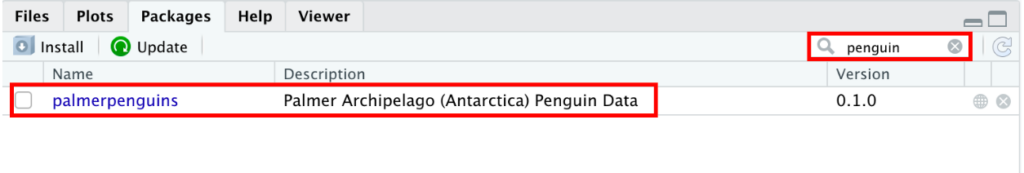 Palmerpenguins package displayed in RStudio Packages tab
Palmerpenguins package displayed in RStudio Packages tab
And, as mentioned before, the palmerpenguins package is not loaded into memory until we call the library function:
library(palmerpenguins)R Package Installation Methods: install.packages vs RScript vs devtools
Understanding when to use each package installation method is crucial for efficient R package management. Here's a comprehensive comparison of the different methods:
Command:
install.packages("packagename")Best for: Installing stable CRAN packages from R console
Advantages: Simple syntax, stable versions, automatic dependency resolution
Disadvantages: Limited to CRAN packages, requires interactive mirror selection
Command:
install.packages("packagename", repos="http://cran.us.r-project.org")Best for: Automated scripts, CI/CD pipelines
Advantages: Non-interactive, reproducible builds
Disadvantages: Requires specifying mirror URL
Command:
Rscript -e "install.packages('packagename')"Best for: Command line automation, server setup
Advantages: No R console needed, fully scriptable
Disadvantages: Requires terminal access
Command:
library(packagename)Best for: Loading already installed packages
Advantages: Loads package functions into memory
Disadvantages: Doesn't install packages, only loads existing ones
Command:
install_github("user/repo")Best for: Installing packages from GitHub
Advantages: Access to latest development versions and beta features
Disadvantages: May be unstable, requires devtools package
Command:
install.packages("file.tar.gz", repos=NULL, type="source")Best for: Development versions, custom builds
Advantages: Latest features, platform-specific optimization
Disadvantages: Requires build tools, more complex setup
How To Uninstall R Packages
We've seen that it is quite easy to install R packages. What about uninstalling them? Well, simple as well. The command to uninstall R packages is remove.packages(). The package's name must be placed between "" as shown in the following example.
Earlier, we installed the palmerpenguins package from GitHub. Using the command below, we can remove this package from R:
remove.packages("palmerpenguins")And the output:
 Removing palmerpenguins package using remove.packages()
Removing palmerpenguins package using remove.packages()
What is Package Masking in R? (How to Fix Function Conflicts)
Some user-contributed R packages may contain functions with the same name as functions in another package. A warning message will pop up in the R terminal when this situation occurs. This situation is called masking.
A function in the same package cannot have two names as well and you cannot create two files with the same name in a directory on your machine. However, functions in different packages can have the same name and do completely different things.
In the above example, we installed the dplyr package loaded the dplyr function in the memory and received objections from three objects (packages) with the same name loaded in the memory.
If you want to use a function that a recently loaded function has masked, you have the following options:
-
Detach the package you don’t use using the
detachfunction, or -
Give the package you want to use a higher priority by loading it before other packages in your project.
To check which package has the highest priority, check the search path:
search()The package with the smaller position number and closer to the GlobalEnv has the highest priority.
In conclusion, user-contributed packages should be used only when you need them. If you plan not to use a package that is loaded in the memory, a good practice is to detach it to avoid further function conflicts.
Remember that when you quit R, all the packages loaded in the memory will be automatically detached. When starting a new session, R will load only the base packages.
Fix R Package Installation Errors (CRAN Mirror, Dependencies, Permissions)
Installing R packages can sometimes fail due to various reasons. Here are the most common errors and their solutions:
Error: "Please select a CRAN mirror for use in this session"
This error occurs when R doesn't know which CRAN mirror to use for downloading packages.
Solution: Specify the CRAN mirror directly in your installation command:
install.packages("packagename", repos="http://cran.us.r-project.org")Or set a default CRAN mirror for your session:
options(repos = c(CRAN = "http://cran.us.r-project.org"))
install.packages("packagename")Error: "Package dependencies not available"
This happens when a package requires other packages that aren't installed on your system.
Solution: Install with dependencies enabled (this is usually the default):
install.packages("packagename", dependencies = TRUE)Error: "Non-zero exit status" or Compilation Errors
These errors typically occur when installing packages from source that contain compiled code.
Solution for Windows: Install Rtools and ensure it's in your system PATH.
Solution for macOS: Install Xcode Command Line Tools:
xcode-select --installSolution for Linux: Install build-essential and development libraries:
sudo apt-get install build-essential libcurl4-openssl-dev libxml2-dev libssl-devError: "Permission denied" or "Cannot install to library"
This occurs when you lack write permissions to the R library directory.
Solution: Install to your user library instead:
install.packages("packagename", lib = Sys.getenv("R_LIBS_USER"))Or on Linux/macOS, use sudo for system-wide installation (not recommended):
sudo Rscript -e "install.packages('packagename')"Error: "Package is not available for this version of R"
Some packages require specific R versions.
Solution: Update R to the latest version, or find a compatible package version:
# Check your R version
R.version.string
# Install specific package version from CRAN archive
packageurl <- "https://cran.r-project.org/src/contrib/Archive/packagename/packagename_version.tar.gz"
install.packages(packageurl, repos=NULL, type="source")Error: "Unable to load devtools" or "install_github not found"
This means the devtools package isn't loaded or installed.
Solution:
# Install devtools if not installed
install.packages("devtools")
# Load devtools into memory
library(devtools)
# Now install_github will work
install_github("user/repo")Timeout Errors During Installation
Large packages may timeout on slow connections.
Solution: Increase the timeout duration:
options(timeout = 300) # 300 seconds = 5 minutes
install.packages("packagename")R Package Management Best Practices (renv, Version Control, Updates)
Following these best practices will help you maintain a clean, efficient R environment and avoid common pitfalls:
1. Use a Package Management Strategy
Create a package installation script for reproducible environments:
# packages.R
required_packages <- c("dplyr", "ggplot2", "tidyr", "readr")
for(pkg in required_packages) {
if(!require(pkg, character.only = TRUE)) {
install.packages(pkg, repos="http://cran.us.r-project.org")
library(pkg, character.only = TRUE)
}
}2. Specify CRAN Mirrors in Scripts
Always specify the CRAN mirror in automated scripts to avoid interactive prompts:
install.packages("packagename", repos="http://cran.us.r-project.org")3. Use renv for Project-Specific Package Management
The renv package creates isolated, reproducible R environments for each project:
# Install renv
install.packages("renv")
# Initialize renv for your project
renv::init()
# Install packages (they'll be project-specific)
install.packages("dplyr")
# Save the state of your project library
renv::snapshot()
# Restore packages on another machine
renv::restore()4. Regularly Update Packages
Keep packages updated for bug fixes and new features:
# Update all packages
update.packages(ask = FALSE, repos="http://cran.us.r-project.org")
# Update specific package
install.packages("packagename", repos="http://cran.us.r-project.org")
# Check which packages have updates available
old.packages()5. Detach Unused Packages
Unload packages you're not using to prevent function masking conflicts and free memory:
# Detach by name
detach(package:packagename, unload = TRUE)
# Detach by position
detach(pos = 2, unload = TRUE)6. Document Package Dependencies
Always document required packages in your R scripts:
# Required packages: dplyr (>= 1.0.0), ggplot2, tidyr
# Install with: install.packages(c("dplyr", "ggplot2", "tidyr"))7. Check Package Installation Paths
Verify where packages are being installed, especially on shared systems:
# Check library paths
.libPaths()
# Install to specific location
install.packages("packagename", lib = "/custom/path/to/library")8. Use Version Control for Package Versions
For reproducibility, consider using exact package versions:
# Install specific version using remotes
install.packages("remotes")
remotes::install_version("dplyr", version = "1.0.7", repos = "http://cran.us.r-project.org")9. Monitor Package Dependencies
Before installing large packages, check their dependencies:
# View package dependencies
tools::package_dependencies("packagename", recursive = TRUE)10. Clean Up Unused Packages
Periodically remove packages you no longer use:
# List all installed packages
installed.packages()[, c("Package", "Version")]
# Remove unused packages
remove.packages("packagename")Frequently Asked Questions
Wrapping Up
Though there are definitely more scenarios to cover, by now you should be fairly confident with how to install R packages from various sources and manage R packages in your system.
Main Commands Summary:
| Action | Command |
|---|---|
| Install from CRAN | install.packages("package") |
| Load into memory | library(package) |
| Unload from memory | detach(package:package) |
| Install from GitHub | devtools::install_github("author/package") |
| Uninstall | remove.packages("package") |
| Check path | .libPaths() |
| Update | update.packages() |
Next Steps:
- Learn how to install R and RStudio
- Explore how to import CSV files in R
If something does not go as planned, you can always refer to the R manuals available by executing help.start() in the R console.